Introduction to iMPI
iMPI database is the first comprehensive resource for iORF-encoded microproteins in tomours. It contains 5,832 intron open reading frames (iORFs) from 4,751 introns with proteomic mass spectrometry evidence. It provides detailed information on these translatable iORFs, including nucleic sequence, peptide sequence, genomic position, coding potential, and information on parent introns, transcripts and genes. Besides, the iMPI database is well annotated provides extended information on iORF-encoded microproteins, such as the post-translational modifications (83,863 phosphorylation sites, 5,141 ubiquitylation sites, 3,574 methylation sites, 3,363 dimethylation sites, 1,996 trimethylation sites, and 735 acetylation sites), subcellular locations, second structures, surface accessibility and three-dimensional structures.
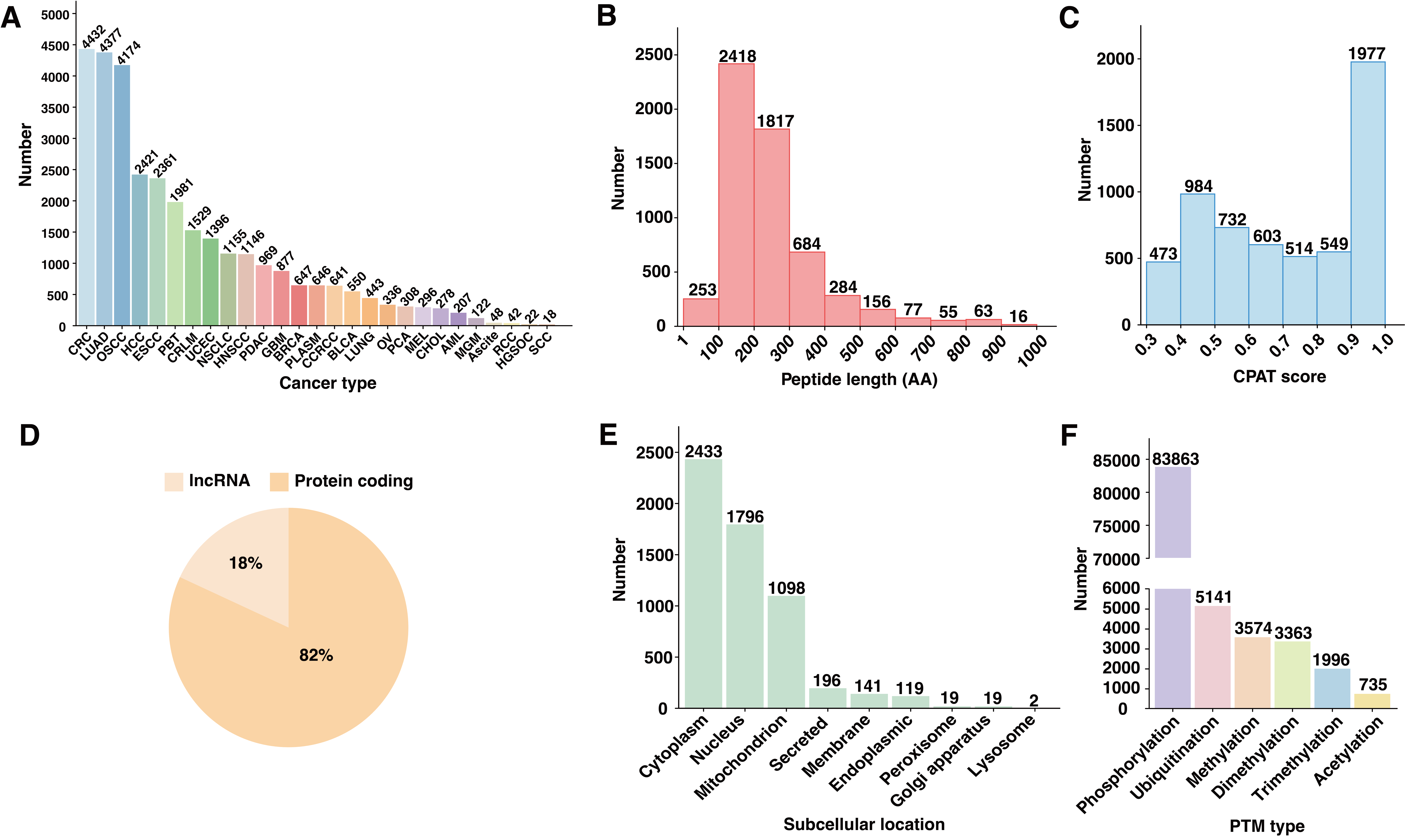
A. Number of IR-encoded peptides with MS evidence across each cancer type. B. Distribution of peptide lengths ranging from 1 to 1000 amino acids (AA). C. Distribution of CPAT score for predicting coding potential of iORFs. D. Gene types of introns. E. Number of IR-encoded peptides localized in different subcellular compartments. F. Number of PTM sites on IR-encoded peptides.
Usage of iMPI
Quick search
On the home page, users can perform keyword-based queries such as gene ID, gene name, transcript ID, and cancer type. To facilitate the search process, examples for each keyword option are provided, including ‘‘ENSG00000107902.14”, ‘‘FLT4”, ‘‘ENST00000370002.8”, and ‘‘LUAD”, which are the gene ID, gene name, transcript ID, and cancer type, respectively.
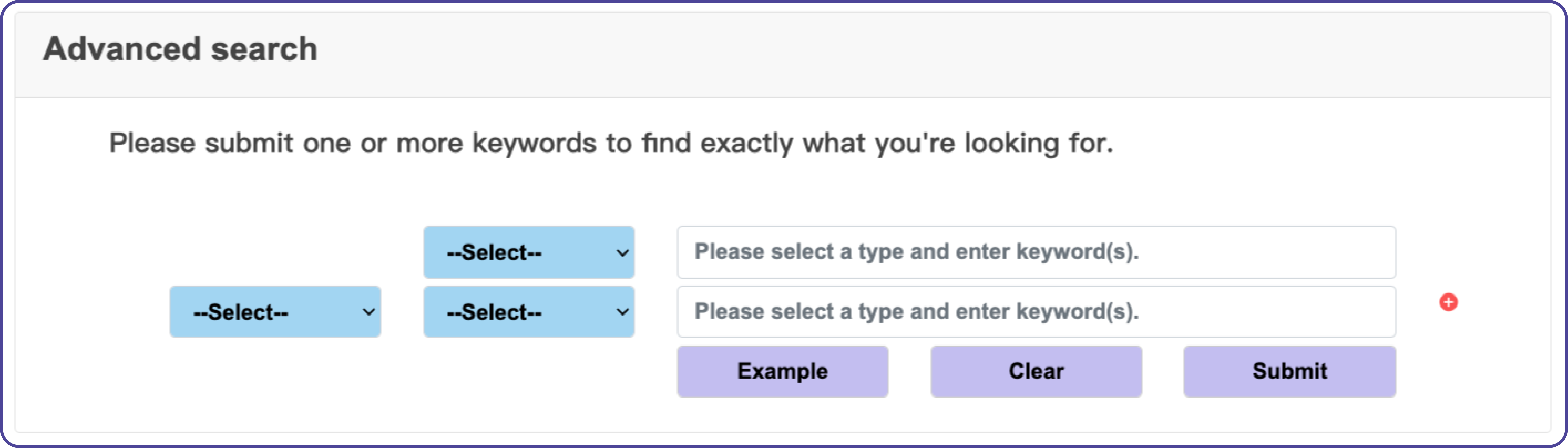
Advanced search
The advanced search function on the “Search” page allows users to submit up to ten keywords and combine them with operators such as “AND,” “OR,” and “NOT” to query IR-encoded microprotein data accurately. An example query like “Gene name = STN1 AND Cancer type = CRC” is available by clicking the “Example” button.
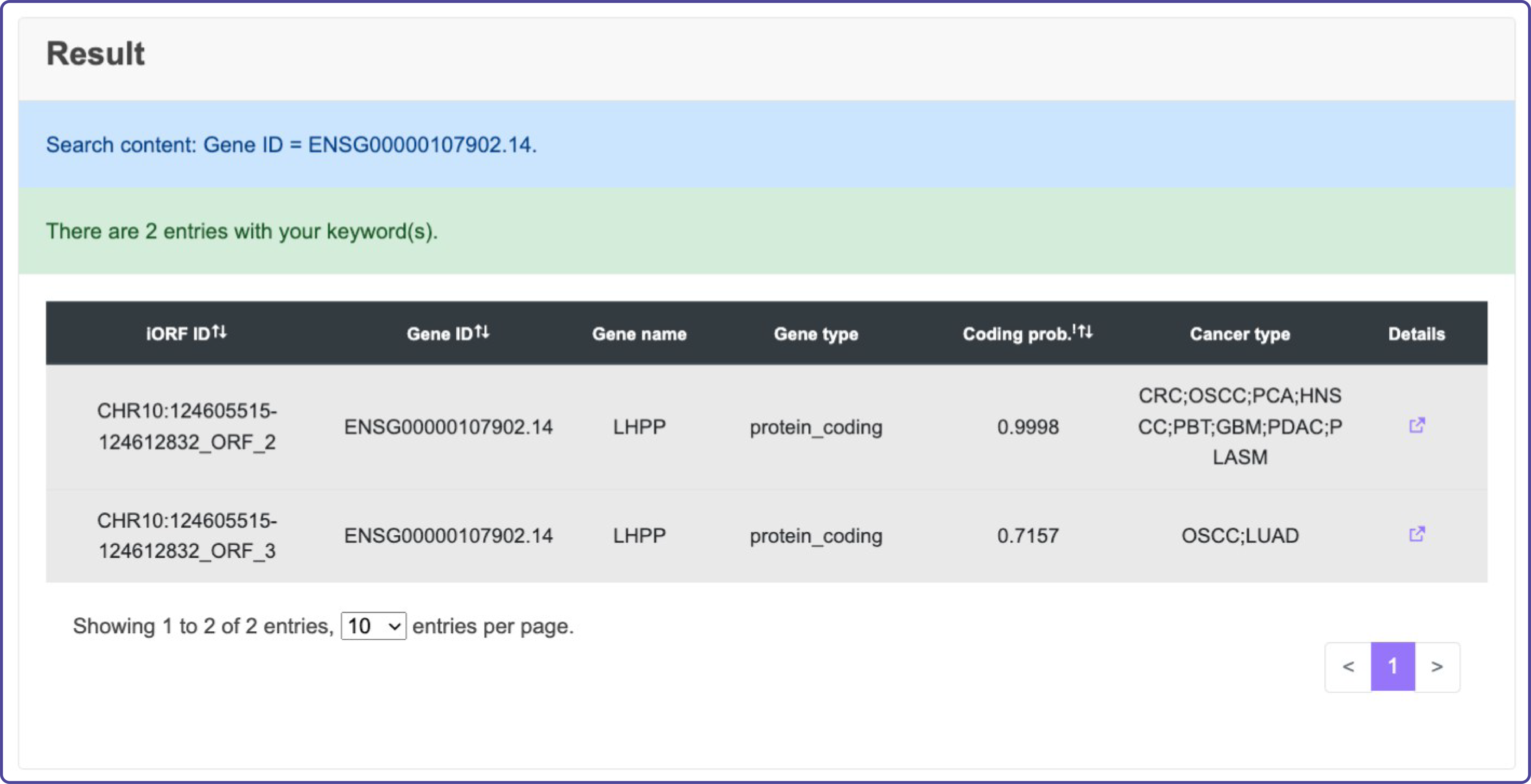
Results
The results are organized in tabular format with columns for iORF ID, gene ID, gene name, gene type, coding probability, and cancer type. The iORF ID represents introns located at different positions on the chromosome. Coding probability indicates the likelihood that an intron encodes a protein, as predicted by the CPAT. Cancer type denotes the presence of protein profile evidence for IR-encoded microproteins in various cancers. Users can view detailed information by clicking the “Details” link.
Details
On the details page, the first section displays intron information, including the iORF sequence, peptide sequence, and intron position, in addition to the data presented in the results table. The database also provides an external link to Ensembl for further information on intron location. The “Coding evidence” section shows MS evidence for peptides encoded by IR. Exploring the subcellular location of proteins is essential for understanding cellular processes. In the iMPI database, the subcellular location of IR-encoded microproteins is displayed in the third section. Furthermore, the PTM information, second structure, surface accessibility, and 3D structure can be visualized in the “Structures” section.

